Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Author affiliation: The Pirbright Institute, Pirbright, UK (A. Di Nardo, A.E. Shaw, J. Wadsworth, K. Parekh, A. Ludi, V. Mioulet, H.M. Hicks, N. Polo, N.J. Knowles, D.P. King); ANSES Laboratory for Animal Health, Maisons-Alfort, France (M. Gondard, G. Girault, C. Bernelin-Cottet, L. Bakkali-Kassimi); Foot and Mouth Disease Institute, Ankara, Turkey (A. Bulut, U. Parlak); Animal Health Institute, Sebeta, Ethiopia (D. Gizaw); Jordan University of Science and Technology, Irbid, Jordan (M. Ababneh); Animal Health Laboratory Directorate, Amman, Jordan (M. Al Ameer); Central Veterinary Laboratories, Baghdad, Iraq (L.M.S. Abdulrasool); Ministry of Municipalities Affairs and Agriculture, Hawrat A’ali, Bahrain (F.S. Al Saloom); Sultan Qaboos University, Muscat, Oman (W.A. Al-Rawahi); Central Laboratory of Animal Health, Muscat (W.A. Al-Rawahi)
Cases of foot-and-mouth disease (FMD) were reported in buffalo in Baghdad, Iraq, during December 2022 (https://wahis.woah.org/#/in-review/4856). Genotyping of FMD virus (FMDV) variable protein (VP) 1 coding sequences generated by the SAP Institute in Turkey identified the causative FMDV as the SAT2/XIV topotype that is present in eastern Africa but is exotic to western Asia (1). Increased surveillance monitored the further spread of this topotype among naive livestock that had not been vaccinated or previously infected with this serotype. By January 2023, fresh outbreaks caused by SAT2/XIV were detected in Bahrain, Jordan, and Oman. In March 2023, outbreaks caused by this topotype were also reported in eastern Anatolia in Turkey and later in central Anatolia and the Adana Provinces. We used whole-genome sequences to investigate the likely timing and route of SAT2/XIV incursion and spread in western Asia.
We performed whole-genome sequencing of 49 SAT2/XIV FMDV isolates from clinical samples submitted to the World Reference Laboratory for FMD (WRLFMD; Pirbright, UK) or the French Agency for Food, Environmental and Occupational Health and Safety (ANSES; Paris, France). Specifically, sequences consisted of 12 samples from Ethiopia collected during May 2022–January 2023, six samples from Iraq collected during December 2022–February 2023, six samples from Jordan collected during January–February 2023, three samples collected in Bahrain in November 2021, seventeen samples from Turkey collected during March–June 2023 (all submitted to WRLFMD), and 5 samples from Oman collected during January–February 2023 (submitted to ANSES) (Appendix Table 1). Sequences were determined at WRLFMD using Illumina MiSeq technology (https://www.illumina.com) as previously described (2) and at ANSES using the Oxford Nanopore MinION platform (https://www.nanoporetech.com) (in-house protocol).
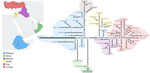
Bayesian phylogenetic reconstruction (3) (Figure 1) demonstrated that the SAT2/XIV FMDV sequences were assigned into distinct sister clades, providing evidence for ancestors circulating in eastern Africa that moved into western Asia causing outbreaks in Iraq, Jordan, and Turkey (most recent common ancestor [MRCA] dated to March 2022 [95% Bayesian credible interval (BCrI) February–May 2022]); and independent introductions into Bahrain (MRCA dated May 2022, 95% BCrI April–June 2022) and Oman (MRCA dated July 2021, 95% BCrI June–September 2021). Those viruses are distantly related (98.70% +13.01 nt identity) to a virus collected during March 2022 from the Wolayita Zone in southwestern Ethiopia. The MRCA of the SAT2/XIV topotype was estimated to be April 1991 (95% BCrI March–May 1991) with an evolutionary rate of 5.46 × 10−3 nt/site/year (95% BCrI 4.686.16 × 10−3 nt/site/year). To our knowledge, SAT2/XIV was detected on only 1 other occasion, in 1991 on a dairy cattle farm located southwest of Addis Ababa, Ethiopia. However, infrequent sampling makes pinpointing the precise source of these viruses within eastern Africa difficult. Statistical parsimony network analysis (4) produced a similar topological representation of the SAT2/XIV FMDV sequences (Figure 2). However, this analysis highlighted several key additional points: infections in Oman were caused by 2 divergent viruses, providing evidence for 2 independent introductions; cases in Bahrain originated from a single virus ancestor, which evolved from viruses circulating in Ethiopia; viruses from Iraq evolved from an eastern Africa ancestor, which also provided the source for onward transmission to Turkey; and the SAT2/XIV outbreaks in Jordan were likely derived from a single independent virus introduction with a different genetic origin than cases reported in Iraq and Turkey.
We observed a single-point mutation at site 8080 (C>T) on the SAT2/TUR/16/2023 genome, shifting the termination to 21 nt downstream of the stop codon at a TGA leading to an extra 7 amino acids (QSLRCHN) on the end of the 3Dpol. A GenBank search of closely related genomes found that this mutation was also present on a SAT2/XIV genome reported separately from Jordan (GenBank accession no. PP112252).
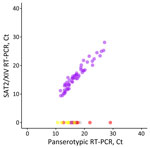
We designed an FMDV lineage–specific real-time reverse transcription PCR (RT-PCR) assay within the 1D region to detect viruses from the SAT2/XIV topotype (Table). We assessed specificity in silico by using alignments of sequences from other viral lineages circulating in the region (A/ASIA/Iran-05, O/ME-SA/PanAsia-2, Asia1/ASIA/Sindh-08, SAT2/VII) and then through experimental testing using representative samples. The real-time RT-PCR correctly categorized 56 samples as SAT2/XIV with no cross-reactivity observed with isolates of serotypes O, A, Asia1, or the SAT1/I and SAT2/VII topotypes (Figure 3).
We obtained evidence of antigen match of SAT2 vaccine strains against SAT2/XIV field viruses using 2D-VN testing (6); all (11/11) isolates were matched to the SAT2 Eritrea 98 vaccine, whereas 8 of the isolates were matched to the SAT2 ZIM 83 vaccine. Although no vaccine-protection data are available for SAT2/XIV viruses, the mean heterologous log10 titers (1.65 + 0.12 for the SAT2 Eritrea 98 vaccine and 1.97 + 0.17 for the SAT2 ZIM 83 vaccine) for bovine serum samples exceeded the cutoff of log10 1.5 previously defined in a SAT 2 potency study for a SAT2/VII isolate (7) (Appendix Table 2).
We report the incursion of the FMDV SAT2/XIV topotype into western Asia and used whole-genome sequencing to reconstruct virus movements from eastern Africa. The emergence of exotic serotype SAT2, and the rapidity with which this virus lineage spread among a naive population, poses threats to the region, as well as to countries in Europe protected by the vaccination buffer zone in Turkish Thrace (8).
FMDV epidemiology is documented by dynamic cross-exchange and long-distance movements of viruses between the 7 endemic pools (9,10). Escapes of FMDV lineages endemic in Africa have been recorded for types A, O, SAT1, and SAT2 viruses; documented examples include SAT1 topotype VI in Bahrain (1962), Greece (1962), Iran (1962–1964), Iraq (1962), Israel (1962), Jordan (1962), Lebanon (1962), Syria (1962), and Turkey (1962–1965) (11). SAT1 was detected on 2 further occasions: an unknown genotype in Kuwait (1969–1970) and Saudi Arabia (1970) (12) and topotype VI in Yemen (1984) (N.J. Knowles, unpub. data). Serotype O, topotype EA-3, was recorded in Yemen during 1971–2009 and more recently in Israel (2017) and Palestine (2017–2018) (13). Serotype A topotype AFRICA lineages have been detected: G-II in Yemen (1985), G-VII in Yemen (1989 and 1998), and G-I in Oman (2018–2021) and Bahrain (2021) (14). Serotype SAT2 lineages have been reported on 6 occasions: topotype IV in Yemen (1990) and Bahrain (2012) and topotype VII in Kuwait (2000), Saudi Arabia (2000), Palestine (2012), and Oman (2015) (15). Those events highlight epidemiologic connections between eastern Africa and western Asia.
Incursions of exotic FMDVs into new areas, and especially from eastern Africa, have been associated with trade in livestock or products of animal origins (10). A recent risk assessment identifies likely pathways of SAT2/XIV diffusion within western Asia, which points to the risk posed by informal livestock trade routes and common grazing (8). Islamic festivals, such as Eid al-Adha, increase demand for meat across the region driving differential pricing, which could also potentially affect the epidemiology of FMD (https://cadmus.eui.eu/handle/1814/75333).
Although data are derived from opportunistic sampling, our findings support 2 hypotheses. First, recent introductions of SAT2/XIV into western Asia are defined by multiple independent incursions; furthermore, the outbreaks detected in Oman and Bahrain appear not to be directly linked, suggesting independent introductions of the virus could have occurred during 2022–2023. Second, cases in Iraq were caused by viruses derived from a single ancestor introduced during late 2022, and the cases subsequently detected in Turkey likely originated from Iraq; however, cases reported in Jordan are likely caused by a virus of a different origin. Uncertainties remain surrounding the origin of SAT2/XIV and how this topotype has been maintained during the 10 years before its reappearance in 2022 in eastern Africa, although a wildlife reservoir, likely within Cape buffalo populations, might represent its ecologic niche.
Dr. Di Nardo is a senior molecular epidemiologist at The Pirbright Institute. His research interests focus on the understanding of processes that drive transmission dynamics of infectious diseases at a multi-scale perspective, integrating analytical methods adopted from molecular evolution, epidemiology, population, and ecosystem ecology.