Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Author affiliation: Center of Infectious Diseases (LUCID), Leiden University Medical Center, Leiden, the Netherlands (J. Gooskens, M.E.M. Kraakman, P. Wörz, S.A. Boers); Leiden University Medical Center, Leiden, the Netherlands (E.H.R. van Essen); Reference Laboratory for Zoonotic Chlamydia Infections, Zuyderland Medical Center, Sittard-Geleen, the Netherlands (E.R. Heddema)
The bacterial genus Chlamydia consists of 14 ubiquitous species that affect a wide range of hosts. The species C. trachomatis, C. pneumoniae, C. psittaci, C. caviae, C. felis, and C. abortus are pathogenic to humans after interhuman or zoonotic transmission. Taxonomy studies have identified a new avian C. abortus subgroup that is an intermediary related to C. psittaci and mammalian C. abortus (1). In 2024, avian C. abortus was associated with human respiratory tract infection and possible human-to-human transmission (2). We report a case of severe community-acquired pneumonia caused by an avian C. abortus genotype not yet associated with human disease.
A previously healthy 74-year-old man from a residential coastal town in the Netherlands was admitted to the hospital during the winter in 2021 with fever, confusion, and progressive dyspnea of 4-day duration. The patient was a nonsmoker and was vaccinated against seasonal influenza and SARS-CoV-2. He lived a socially withdrawn lifestyle and had no exposure to ruminants or domestic birds, although he regularly fed wild aquatic birds during the winter months. He reported noteworthy exposure to wild birds, including seabirds, which included hand feeding and occasional contact with bird droppings on his clothing.
At hospital admission, a physical examination revealed a body temperature of 39.3°C, pulse of 162 beats/min, blood pressure of 127/77 mm Hg, and respiration rate of 42 breaths/min. Laboratory results showed an unremarkable leukocyte count of 7.49 × 109 cells/L (reference range 4.0–10.0 × 109 cells/L), acute lymphocytopenia of 0.43 × 109 cells/L (reference range 1.0–3.5 × 109 cells/L), unremarkable serum creatinine level of 101 µmol/L (reference range 64–104 µmol/L), and elevated C-reactive protein level of 305 mg/L (reference range <5.0 mg/L). Other findings included hyponatremia with a serum sodium level of 129 mmol/L (reference range 136–145 mmol/L) and elevated levels of creatine kinase, 531 U/L (reference range <171 U/L); plasma fibrinogen, 7.3 g/L (reference range 2.1–3.8 g/L); serum lactate dehydrogenase, 397 U/L (reference range <248 U/L); blood urea, 11.7 mmol/L (reference range 2.5–7.5 mmol/L); serum glucose, 10 mmol/L (reference range 3.9–7.7 mmol/L); γ-glutamyl transferase, 71 U/L (reference range <55 U/L); aspartate aminotransferase, of 87 U/L (reference range <35 U/L); and cardiac troponin T, 58 ng/L (reference range <14 ng/L). All other laboratory findings were unremarkable.
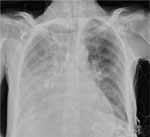
The patient was transferred to the intensive care unit because of hypoxemic respiratory failure (SaO2 <90%) and progressive pulmonary consolidations with pleural effusion (Figure 1) requiring invasive mechanical ventilation. Blood and sputum cultures collected before intravenous cefuroxime and ciprofloxacin empirical therapy showed no microbial pathogens. Bronchial aspirate obtained by fiberoptic bronchoscopy was negative for respiratory viruses, Legionella spp., Mycoplasma pneumoniae, and Chlamydia pneumoniae by real-time PCR; however, B-CAP real-time PCR (Biolegio, https://www.biolegio.com) detected Chlamydia DNA. Quantitative 16S micelle PCR and next-generation sequencing–based (3) and metagenomic-based (4,5) microbiota profiling showed a high relative abundance of Chlamydia spp. in the lower respiratory tract; levels were >100 million 16S rRNA gene copies per milliliter of bronchial aspirate (Appendix Figure 1). Metagenomic profiling suggested a high abundance of Chlamydia spp. intermediaries related to C. psittaci and C. abortus. The reference laboratory detected C. abortus carrying a plasmid inherent in avian C. abortus (6). Multilocus sequence typing revealed sequence type 359. Phylogenetic analyses of concatenated multilocus sequence typing and plasmid II xerC gene sequences confirmed avian C. abortus (Figure 2) (6,7). Chlamydial outer membrane protein A gene sequencing identified a single Chlamydia spp. matching provisional genotype R54 (Appendix Figure 2), previously isolated from migratory seabirds in polar regions but never from humans or other mammals (8,9). The avian C. abortus clinical isolate KML-2021 (GenBank accession nos. OR665720 and PQ001575) obtained from the patient in this study was genetically different from avian C. abortus clinical strain NL2335-4C (GenBank accession nos. CP158097 and CP158098) and related clinical isolates or strains from other patients in the Netherlands (Figure 2; Appendix Figure 2) (2).
The patient’s respiratory condition improved with doxycycline treatment directed at Chlamydia spp. At 1-year clinical follow-up, the patient noted no recurrences. The patient provided written informed consent for publication to the treating physician (E.H.R.v.E.) after the 1-year clinical follow-up. All authors confirmed that subject protection guidelines and regulations were strictly followed. We notified the Public Health Service for the Hollands Midden region in Leiden, the Netherlands, of the zoonotic Chlamydia spp. infection and did not identify any human-to-human transmission events.
In conclusion, our findings confirm the zoonotic potential of avian C. abortus to cause severe community-acquired pneumonia in humans. Increased awareness is warranted to establish the occurrence, clinical manifestations, and global geographic distribution of that rare zoonotic disease. We recommend molecular surveillance studies in wild and captive birds to evaluate sources of contamination of different avian C. abortus genotypes.
Dr. Gooskens is a clinical microbiologist at the Department of Medical Microbiology, Leiden University Medical Center, Leiden, the Netherlands. His research interests focus on zoonotic Chlamydia spp. infections and genomic clinical surveillance of CC398 Staphylococcus aureus.