Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Author affiliation: Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA (C.M. George, K. Endres, J. Perin, D.A. Sack); Université Catholique de Bukavu, Bukavu, Democratic Republic of the Congo (A. Namunesha, W. Felicien, P. Sanvura, J.-C. Bisimwa, G. Maheshe, C. Cikomola, L. Bisimwa, A. Mwishingo); Ministère de la Santé, Bukavu (J. Bengehya); University of New Mexico Health Sciences Center, Albuquerque, New Mexico, USA (D. Domman)
An estimated 2.9 million cholera cases and 95,000 deaths occur annually in cholera-endemic countries (1). The Democratic Republic of the Congo (DRC) has one of the highest rates of cholera in Africa (2). In 2017, the Global Task Force on Cholera Control released the document Ending Cholera: A Global Roadmap to 2030 to reduce cholera deaths by 90% in the DRC and eliminate cholera in 20 other countries by 2030 (3). To address cholera in transmission hotspots, the task force recommends oral cholera vaccine (OCV) campaigns and case area–targeted water, sanitation, and hygiene (WASH) interventions should be implemented in a ring around cholera cases. However, a global OCV stockpile shortage has necessitated identifying the duration of protection conferred by OCVs to determine the optimal frequency for OCV campaigns (4). Most studies on OCV effectiveness have been from India and Bangladesh and have evaluated the effectiveness of 2-dose OCV protection (H. Xu et al., unpub. data, https://doi.org/10.1101/2024.08.13.24311930). Only 4 studies have evaluated single-dose OCV vaccine effectiveness (2 of those for a duration of >24 months), and all except 1 used the Shanchol vaccine, which is no longer produced (H. Xu et al., unpub. data).
Few studies have combined evaluations of OCV effectiveness and genomic surveillance of clinical and environmental Vibrio cholerae isolates by using whole-genome sequencing (WGS), which can provide valuable information on how OCV campaigns affect V. cholerae transmission dynamics and can identify genetic characteristics of circulating V. cholerae strains. WGS is also a valuable tool to link cholera epidemics globally and investigate V. cholerae transmission by distinguishing strains on the basis of single-nucleotide polymorphisms (SNPs) (5). Genomic data from 45 countries in Africa revealed that the V. cholerae seventh pandemic El Tor (7PET) strain was introduced into Africa >15 times since 1970 (6). Previous genomic studies (2018–2024) in DRC have found V. cholerae 7PET strains belong to clades AFR10d, AFR10e, and ARFR10w (7,8).
We previously used WGS to analyze water and clinical sources of V. cholerae collected from patient households to investigate cholera transmission dynamics in Bangladesh (9). We found that 80% of cholera patient households had isolates from water that were closely related to clinical isolates; 20% of households had clinical isolates from infected persons that were more closely related to clinical isolates from other households than to source water isolates in their own household. Those results were consistent with person-to-person and water-to-person V. cholerae transmission. Genomic studies to elucidate transmission dynamics of V. cholerae infection in cholera patient households in sub-Saharan Africa are lacking; previous studies have been exclusively in South Asia.
We conducted 4 years of epidemiologic and genomic surveillance of V. cholerae in eastern DRC to achieve the following aims. First, we evaluated the effectiveness of a single-dose of Euvichol-Plus (EuBiologics, http://eubiologics.com), a killed whole-cell OCV (kOCV), during the 24-month period after a preventive kOCV campaign. Second, we investigated V. cholerae transmission dynamics among cholera patients, household members, and water sources by using WGS. Third, we determined the spatiotemporal spread of V. cholerae in this cholera-hyperendemic region.
Ethics approval
We conducted this study in urban Bukavu, South Kivu Province, DRC. We received ethics approval for this study from Johns Hopkins Bloomberg School of Public Health, (Baltimore, Maryland, USA) and Catholic University of Bukavu (Bukavu, DRC). All participants or their guardians provided written informed consent.
Study Design and Protocol
During March 2020–March 2024, we conducted passive cholera surveillance at 115 healthcare facilities in Bukavu. Patients with diarrhea who were admitted during this time had their feces tested for V. cholerae by using bacterial culture. We defined cholera patients as patients with diarrhea who had a V. cholerae–positive fecal sample by bacterial culture. We conducted a prospective cohort study of household contacts of cholera patients during December 2021–December 2023 to investigate cholera transmission dynamics within cholera patients’ households. We defined household contacts of cholera patients as those sharing a cooking pot and residing in the same home with the cholera patient for the previous 3 days. We enrolled household contacts in the study within 24 hours of enrolling the corresponding cholera patient. We determined the sample size for the prospective cohort study by using the number of cholera patients who could be screened and who were willing to participate in the cohort study. We visited cholera patient households on days 1, 3, 5, 7, 9, and 11 (visits 1–6) after the household’s index cholera patient was admitted at a health facility to conduct clinical surveillance. For clinical surveillance, we collected a fecal sample from the cholera patient and household contacts during each household visit to test for V. cholerae by using bacterial culture. We conducted an unannounced spot check at each timepoint to collect a sample of the household’s water source and stored drinking water to test for V. cholerae by bacterial culture.
During December 28, 2021–January 2, 2022, and March 31–April 4, 2022, the DRC Ministry of Health delivered 1.04 million doses of Euvichol-Plus kOCV in Bukavu as a preventive OCV campaign. Vaccines were delivered through a combination of door-to-door visits and designated healthcare facilities. We assessed OCV vaccination status for patients with diarrhea and their household members through self or caregiver reporting during the time of patient treatment in the healthcare facility or during a home visit conducted the same or the following day. Study research officers administered a structured questionnaire, which obtained information on OCV administration and the date and number of doses. An OCV vaccine card was shown, along with a photo of the person consuming OCV. We defined informal settlements as areas where no household connections to piped water existed.
Laboratory Analyses
All whole fecal samples were brought to the Preventative Intervention for Cholera for 7 Days program Enteric Disease Microbiology Laboratory in Bukavu within 3 hours of the sample being produced, and water samples were brought to the laboratory within 3 hours of collection for V. cholerae analysis by bacterial culture as previously described (10). We preserved isolated bacteria as stabs in nutrient agar or on Whatman filter paper to preserve bacterial DNA.
WGS
We extracted DNA from isolates preserved as agar stabs by using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research, https://www.zymoresearch.com) and extracted genomic bacterial DNA from filter paper by using published Chelex methods (11). The SeqCenter (https://www.seqcenter.com) and University of New Mexico Health Sciences Center performed WGS. We processed short reads by using the Bactopia pipeline (12) and SPAdes version 3.10.0 (13) and annotated by using Prokka version 1.520 (14). We performed genome completeness estimates and checks for contamination by using CheckM version 1.0.722 and Kraken version 0.10.6 (15,16). We deposited all next-generation sequencing data from this study under the National Center for Biotechnology Information BioProject database (https://www.ncbi.nlm.nih.gov/bioproject; no. PRJNA1210607).
Genomic and Phylogenetic Analyses
We mapped paired-end reads for 255 7PET isolates and compared variants with the V. cholerae O1 El Tor reference genome N16961 (GenBank accession nos. LT907989 and LT907990) by using snippy version 4.6.0 (https://github.com/tseemann/snippy) via the Bactopia pipeline (12) to generate a reference-based alignment containing 329 variable SNP sites. All 255 isolates mapped with >90% of the reference genome; thus, we further analyzed all 255 isolates. We generated a pairwise SNP matrix for the 255 7PET isolates by using pairsnp (https://github.com/gtonkinhill/pairsnp). We used ARIBA version 2.14.7 (https://github.com/sanger-pathogens/ariba) to determine mutations in the wbeT gene by comparing sequences with a reference wild-type Ogawa wbeT gene from the European Nucleotide Archive database (https://www.ebi.ac.uk/ena; accession no. AEN80191.1)
We compared all 7PET isolates from this study with 1,418 globally representative 7PET strains to assess phylogeographic relatedness (17). We used the 12,561 variable site (SNPs) alignment to build a maximum-likelihood phylogeny; we used IQ-Tree version 1.6.12 and the general time reversible substitution model with the gamma distribution to model site heterogeneity with 10,000 ultrafast bootstraps and used 10,000 bootstraps for the Shimodaira-Hasegawa–like approximate-likelihood ratio tests (18). We visualized phylogenies by using ggtree version 1.6.11 (19) and rooted the trees by using the pre-seventh pandemic V. cholerae strain M66.
Local Transmission Analyses
We gained insights into local transmission dynamics from a 329 SNP reference-based alignment of the 255 7PET isolates. We built a maximum-likelihood tree by using IQ-Tree and the models and bootstraps as described previously. We visualized phylogenies by using Microreact (20). We created minimum spanning trees by using GrapeTree version 1.5.0 (21) and visualized geospatial data for cases by using Python geopandas (https://github.com/geopandas/geopandas) and contextily (https://github.com/geopandas/contextily) packages.
Statistical Analysis
We assessed single-dose Euvichol-Plus kOCV effectiveness during the 24-month period after vaccination by using a test-negative design among patients with diarrhea. We used logistic regression models with cholera infection as the outcome (defined as a positive V. cholerae bacterial culture) and kOCV vaccination status (whether patients with diarrhea reported receiving 1 dose of kOCV during the kOCV campaign period) as the predictor. In this model, the odds ratio was the odds of single-dose kOCV vaccination effectiveness in the case-patients (cholera-positive patients with diarrhea) compared with controls (noncholera patients with diarrhea). We calculated vaccine effectiveness by using the equation (1 – OR) × 100%, and we excluded persons who reported receiving 2 doses of kOCV from the analysis. We performed analyses by using SAS version 9.4 (SAS Institute Inc., https://www.sas.com). We performed permutation tests by using R (The R Project for Statistical Computing, https://www.r-project.org) and Python to analyze pairwise comparisons of genomic data. For pairwise comparisons, we compared SNP counts for each strain with those from the V. cholerae reference strain N16961.
Epidemiology
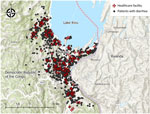
During March 2020–March 2024, we identified V. cholerae in fecal samples from 342 (30%) of 1,154 patients with diarrhea via bacterial culture (Appendix Figure 1). We mapped the 115 healthcare facilities and households of patients with diarrhea surveilled in this study (Figure 1). We observed an annual bimodal peak of cholera during the dry (June–August) and rainy (September–January) seasons in 2020 and 2023 (Appendix Figure 2). In December 2021, a kOCV campaign distributed 1.04 million doses of Euvichol-Plus within Bukavu (study surveillance site). After that kOCV campaign, sporadic cholera patients were observed through January 2023. Then, ≈14 months after the kOCV campaign (June–November 2023), a large cholera outbreak occurred in this same area. Nine percent (309/3,395) of persons residing in informal settlements within the study area reported receiving >1 kOCV dose during December 2021–April 2022. During the 4-year surveillance period, stored and source water samples were collected from 177 cholera patient households; 9% (16/172) of households had V. cholerae–positive stored water samples and 5% (8/151) of households had positive source water samples. A total of 29 water samples were V. cholerae positive over the study period (Appendix Figure 3). During October 2021–November 2022 (including the 12-month period after the kOCV campaign was initiated), no V. cholerae was detected in drinking water samples, despite the presence of culture-confirmed cholera patients.
OCV Vaccine Effectiveness
Surveillance healthcare facilities admitted 750 patients with diarrhea during the 24-month period after the kOCV campaign (December 2021–December 2023). We recorded demographic characteristics for kOCV vaccinated and unvaccinated persons (Table). During the 3 nights before hospitalization, 94% (708/750) of patients with diarrhea reported residing in their current residences; 12% (93/748) had running water inside their home. Twelve percent (93/750) (15 cholera and 78 noncholera patients with diarrhea) of patients reported receiving >1 dose of kOCV during December 2021–April 2022; only 2% (14/750) of patients reported receiving 2 kOCV doses, and 13% (12/93) of kOCV-vaccinated patients showed a vaccination card. During this period, 208 (193 unvaccinated and 15 kOCV vaccinated) patients had cholera; 531 patients with diarrhea were >1 year of age. The unadjusted single-dose kOCV vaccine effectiveness in the first 24 months after vaccination was 59.8% (95% CI 19.7%–79.9%) for persons >1 year of age and, after adjustment for age (continuous variable), was 58.7% (95% CI 17.3%–79.4%). We also calculated the single-dose kOCV vaccine effectiveness according to time interval (first 12-month and second 12-month period after vaccination) for persons >1 year of age (Appendix Table). We excluded 10 patients with diarrhea from this analysis because they received 2 doses of kOCV.
Genomic Analyses
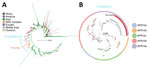
We sequenced 255 V. cholerae 7PET genomes from samples collected during December 2020–December 2023 within the study area. Of those genomes, 247 were clinical isolates (158 Ogawa and 89 Inaba serotypes) from the fecal samples of 243 persons residing in 200 households (198 patients and 45 household contacts), and 8 were from water samples. The WGS analysis included 75% (255/342) of cholera patient households during the surveillance period, and all available isolates were sequenced. To investigate the genetic relatedness of the V. cholerae isolates, we analyzed the pairwise SNP differences across 255 genomes. Of the 200 households in the study, 31 households were represented by >1 sample, 26 households had >1 study participant with samples, and 5 households had both clinical and water V. cholerae isolates. Maximum-likelihood phylogenic analyses of the 255 7PET V. cholerae genomes indicated the V. cholerae isolated from both clinical and water samples were closely related, and limited genetic divergence occurred over the study period (Figure 2, panels A, B). Among all isolates, the minimum number of SNP differences was 0 and the maximum number was 65 (median 13) (Figure 3, panel A). For isolates from the same person, the range was 0–1 SNP and median 0 SNPs. For isolates from the same household (isolated from both clinical and water samples), the pairwise differences range was 0–14 SNPs (median 0) (Figure 3, panel B). We found a significant difference in the median number of SNPs among isolates from different households compared with those from the same household (p<0.0001 by Mann-Whitney U test). In the 3 of 5 households with both clinical and water V. cholerae isolates, the isolates had 0 SNP differences among them; the other 2 households had 1–3 SNP differences between the clinical and water samples. This result provides evidence that the same strain causing infections in household members was also detected in the household water. However, we cannot determine whether the water was the source of infection or whether the water was contaminated after an infection occurred in a household member. The median difference in the number of SNPs was significantly lower for isolates from cholera patients before the kOCV vaccine campaign (median 6 [range 0–58]) compared with the number of SNPs after the kOCV vaccine campaign (median 12 [range 0–31]) (p<0.0001 by Mann-Whitney U test).
To determine how the 255 V. cholerae isolates from this study fit within the larger diversity of 7PET strains, we contextualized those genomes within a collection of 1,422 additional 7PET genomes (Figure 4) (17). Phylogenetic analysis placed the 255 isolates within the T10 lineage of the AFR10e clade. Our isolates, sampled during 2020–2023, were similar to other recent samples (2015–2020) from DRC in the same region of South Kivu (8). Genomic analysis of the wbeT gene indicated that samples from 2020–2022 had intact wild-type wbeT genes, which indicates an Ogawa serotype; we observed 1 exception for a sample from 2020 with a frameshift mutation (N165fs), which would likely result in an Inaba serotype. However, all samples from 2023 after the OCV campaign appear to have a fragmented wbeT gene marked by insertion of mobile elements, likely conferring an Inaba serotype. In line with other AFR10e strains, our isolates harbored mutations in gyrA (S83I) and parC (S85L) genes, which have been proposed to reduce susceptibility to fluoroquinolones (8).
In the urban cholera-endemic setting in DRC where we conducted our study, annual bimodal peaks of V. cholerae clade AFR10e infections corresponded with the dry and rainy seasons. One third of patients with diarrhea attending 115 surveillance healthcare facilities were confirmed to have cholera by bacterial culture; 9% of stored water and 5% of source water samples from cholera patient households contained V. cholerae. This finding suggests that both source water and contamination of stored water might be potential transmission routes for V. cholerae infections in cholera patient households. A large clonal cholera outbreak occurred 14 months after >1 million doses of Euvichol-Plus kOCV vaccines were distributed in the same area. This large outbreak occurred despite vaccine campaign, possibly because of the low (9%) kOCV coverage within informal settlements in Bukavu, which are often hotspots for cholera because of limited access to improved drinking water sources, sanitation options, and poor hygienic conditions (22). No water samples were positive for V. cholerae during the 12-month period after the kOCV campaign was initiated, despite the presence of cholera patients. Future studies should investigate the effect of kOCV campaigns on V. cholerae persistence in the environment.
Using a test-negative design, we found that a single dose of Euvichol-Plus provided modest protection against medically attended cholera during the 24 months after vaccination, consistent with findings from another study (H. Xu et al., unpub. data). This test-negative design to evaluate kOCV effectiveness could be incorporated into existing cholera surveillance activities globally and could be integrated as part of kOCV vaccine campaign rollouts. Our findings suggest that, with increased vaccination coverage in informal settlements, single-dose kOCV campaigns are a promising approach to deliver vaccines along with WASH programs to reduce cholera in the DRC.
Our genomic findings highlight several points. First, we did not detect any samples that grouped in the V. cholerae clade AFR10d, a finding that seems to further corroborate that clade AFR10e replaced AFR10d around 2018 in this region (23). Second, during the subsequent cholera outbreaks after the large kOCV campaign, we observed a change in V. cholerae serotype from Ogawa to Inaba, which was likely related to a fragmented wbeT gene marked by insertion of mobile elements. This finding is consistent with another study that observed a serotype switch after a vaccination campaign (24). In addition, we observed an increase in the number of SNP differences among V. cholerae isolates collected after the kOCV campaign, compared with those collected before the campaign. However, both our findings and those from the previous study are observational, and causality cannot be inferred. Third, this study provided evidence that household water contained the same isolates that caused cholera infections among household members. This result combined with the finding that clinical isolates in the same household were more closely related than isolates from different households (even when no V. cholerae was found in household drinking water) suggests a combination of person-to-person and water-to-person cholera transmission. Our findings suggest it is critical for WASH programs to place emphasis on both chlorination of household water and handwashing with soap to prevent cholera transmission.
The first strength of our study is the 4-year duration of epidemiologic and genomic surveillance of cholera patients and their water sources before and after a kOCV campaign. Previous longitudinal studies including both clinical and environmental surveillance have been almost exclusively performed in South Asia. Second, we evaluated the effectiveness of a single-dose of Euvichol-Plus over a 24-month period, building on previous studies that focused mostly on 2-doses of kOCV and evaluated the Shanchol kOCV, which is no longer produced. One study evaluated the single-dose kOCV effectiveness of Euvichol-Plus for a period of >24 months in DRC and found 54% effectiveness 12–17 months after vaccination, similar to our study (25). In addition, the test-negative design for kOCV effectiveness builds on observational studies using community controls by reducing the likelihood of differences in care-seeking behavior between V. cholerae–infected persons and controls, which can introduce substantial bias in observational studies of vaccine effectiveness. Finally, the inclusion of the genomics data enabled us to investigate how water and clinical V. cholerae strains evolved over time before and after a kOCV campaign and investigate V. cholerae transmission dynamics in cholera patient households.
The first limitation of our study is that our surveillance focused only on an urban setting; therefore, we cannot generalize our findings to rural settings. Additional data on single-dose kOCV vaccine effectiveness is needed in rural settings globally. Second, as with all observational study designs, misclassification of a patient’s vaccination status and cholera infection outcome is possible. Also, the vaccine campaign in this study was a preventive campaign in an area identified as a hotspot; however, because of a global kOCV vaccine shortage, all campaigns are reactive and respond to outbreaks. Future studies are needed to evaluate single-dose kOCV effectiveness during reactive kOCV campaigns worldwide.
In conclusion, we observed annual bimodal peaks of V. cholerae clade AFR10e in an urban cholera-endemic area in eastern DRC. One third of patients with diarrhea admitted to surveillance healthcare facilities were confirmed to have cholera by bacterial culture. A large clonal cholera outbreak occurred 14 months after a single dose kOCV campaign, and the severity of the outbreak might have been related to the low vaccine coverage in informal settlements. Genomic analyses suggest that both person-to-person and water-to-person transmission occurred. A single-dose of Euvichol-Plus kOCV provided modest protection against medically attended cholera during the 24 months after vaccination. Our findings indicate that single-dose kOCV campaigns in combination with WASH programs should be used to reduce disease in cholera-endemic settings worldwide.
Dr. George is a professor and infectious disease epidemiologist. Her research focuses on developing and evaluating community and healthcare facility–based infectious disease control programs to reduce infections in low- and middle-income countries and low-resource settings globally.
We thank the study participants for their support in implementing this study, our funders, and our research officers Raissa Boroto, Freddy Mwambusa, Pacifique Kitumaini, Blessing Muderhwa, Jean Claude Lunyelunye, Feza Rugusha, Gisele Kasanziki, Brigitte Munyerenkana, Jessy Tumsifu, Pascal Kitumaini, Emmanuel Buhendwa, and Julienne Rushago, who played a crucial role in the success of this work.
This work was funded by Wellcome and UK aid from the Foreign and Commonwealth Development Office (grant nos. 215674Z19Z and 1R01AI148332-01 to C.M.G.) and by the National Institutes of Health (grant nos UL1TR001449 and KL2TR001448 to D.D; 2R01AI123422-06A1 to D.A.S.).
The funding agencies were not involved in study design, data collection, data analysis, and data interpretation, or manuscript submission. The views expressed do not necessarily reflect the official policies or views of the Foreign and Commonwealth Development Office.